Research topics
Chemical Biology is the field of using and developing chemical techniques in order to study biology. In the past, we developed new rapid bioconjugation reactions and researched new ways to selectively modify DNA or RNA nucleobases. While these projects continue, they have also led us into new areas of chemical biology and translational research such as DNA encoded libraries and chemical protein silencing.
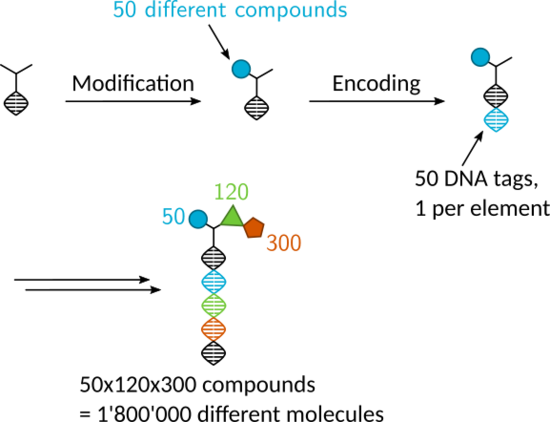
DNA encoded libraries
Target based drug discovery usually starts with high throughput screening: thousands of compounds are tested individually against the desired target. Although this process is usually automated, it is still expensive in terms of time and space since each compound needs to be stored and tested separately. DNA encoded chemical libraries (DECL) comprise a pooled collection of compounds where each individual is covalently linked with a unique DNA barcode. This association of phenotype (compound) and genotype (DNA barcode) allows the parallel screening of millions of compounds against a protein target in the same mixture. With the help of PCR and DNA sequencing, the barcode can then be read out, enabling the determination of binders for that protein. The simple phase-based purifications of DNA chemistry, as in solid-phase synthesis, facilitates split-and-pool-synthesis, making the generation of a diverse combinatorial libraries possible.
In our research group, we strive to synthesize libraries with unique scaffolds involving novel reactions. We also try to innovate selection methods to improve the final data quality in encoded library selections.

Chemical protein silencing
The treatment of diseases with drugs usually involve a small molecule that binds and inhibits a disease-relevant protein. A new class of molecules have recently been discoverd that can bind and destroy and proteins. Chemical protein silencing functions by hijacking the natural cellular mechanism for protein turnover known as the ubiquitin proteasome pathway. The transfer of ubiquitin chains to proteins is often a signal for destruction in the protein wood-chipper known as the 26S proteasome. E3 ligases are the masters regulators of this process because they control the coupling of ubiquitin to substrate proteins. Recent work has shown that small molecules can sometimes bind and change the substrate specficity of E3 ligases, coaxing them into degrading new substrates. This exciting innovation has opened the door to building small molecule protein degradation catalysts that perform the same sort of function as siRNA, but with all the advantages of small molecules.
Our group tries to find new E3 ligases that can be used for targeted degradation of proteins or other cellular processes that we could hijack to degrade specific proteins.

