/ News, Research, Doctorate/PhD, Publication
Expanding the DEL toolset: A functional approach to DEL selections
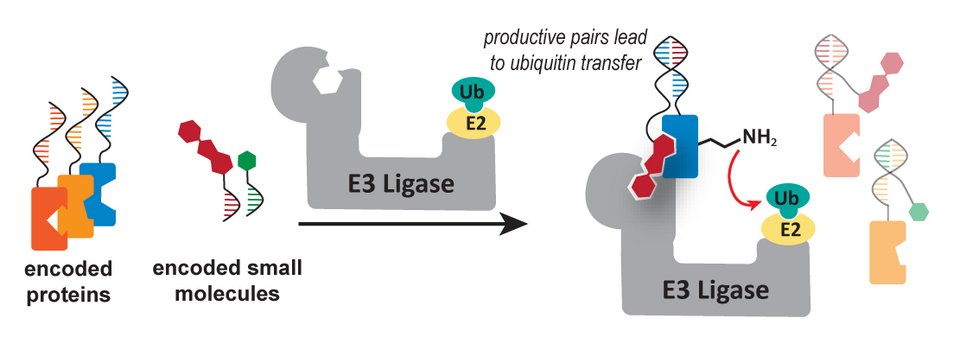
While normal DELs find hits based on their ability to bind a molecule, this new approach sets the foundation to search specifically for molecules that cause ubiquitin transfer from CRBN-E3-Ligase to a POI.
In a typical DNA encoded library (DEL) selection, a protein of interest (POI) is immobilized in a solid support and is incubated with a DEL. The supernatant than gets removed, and the POI on the solid support gets washed a few times, and finally, protein gets denatured to elute DEL member's that bind the protein. These steps can be repeated a 2-3 times, always using the output of the previous selection round as the new DEL input, to improve the ratio of true binders to random ones. This approach works if you search for binders of a specific POI - but they cannot deliver molecules that guarantuee a function.
Molecules that cause one protein to be bind to another one are called, depending on their size and properties, molecular glues. A certain subset of molecular glues got widely known as chemical protein silencer, as they bring a protein close to an E3 ligase. This ligase complex transfers ubiquitin onto that protein, which marks its for degradation. An (in)famous exampel of this is the small molecule thalidomide, also known as Contergan. The exact mechanism on how these birth defects was only discovered in recent years. The molecule, thalidomide, binds the E3 ligase cereblon. This modifies the surface of cereblon (CRBN) and causes a range of effects which are teratogenic. However, it also causes the degradation e transcription factors ikaros (IKZF1) and aiolos (IKZF3) which can be used to fight cancer cells.
As our research group is involved in both of these hot research areas, it was only natural for us to think of a strategy which can be used to have DEL selections for functional ubiquitin transfer, which is the first step to find new molecular degraders. And in this paper, Dr. Pinwen Cai presents the possible strategy to do exactly that.
The system was inspired by the DNA-templated approach from David Liu. One DNA strand is bound to the POI (in our case, the degron of IKZF1 expressed as a fusion to SNAP-Tag - a protein that enables the conjugation of Benzylguanosine-derivatives to itself). The DNA has an encoded binding site, on which a second DNA strand can selectively anneal to. This DNA is bound to a thalidomide-derivative (pomalidomide), which brings the POI and the molecule in proximity. Now if this DNA construct is incubated with the (commercially available) ubiquitination system in presence of HA-tagged Ubiquitin, the small molecule catalyzes the transfer of Ubiquitin onto the POI - only if complex of all three partners actually forms. The HA-Ubiquitin-Tagged DNA strand can then be enriched for by incubating the system with anti-HA beads and running selective qPCR afterwards to quantify the strands.
The paper was published in Chemical Science and is the first peer-reviewed ubiquitin-transfer focused DEL selection method. It marks the end of a successful PhD for Dr. Pinwen Cai—and the start of a new era in functional DEL selections. Chiara Disraeli continues to work on this system. Be tuned for more advances in these topics.
