/ News, Research, Publication
A new method to select hits in DNA encoded libraries
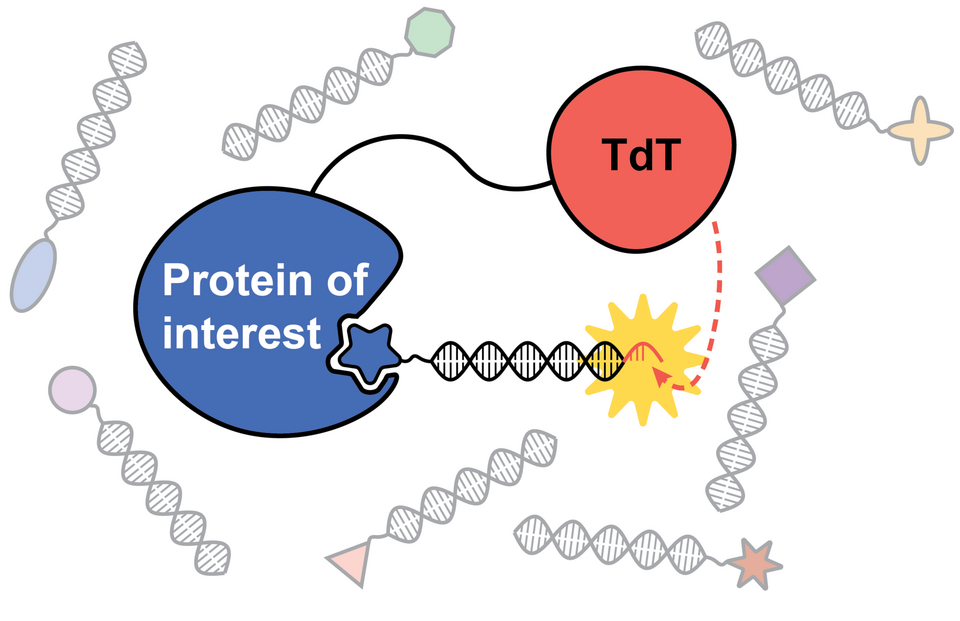
Our very first JACS paper is about DNA encoded libraries. It describes a novel method to select for hits that performs significantly better than affinity selections - the industry standard (so far). And the best of it - its compatible to any library design, as long as the DNA has a free 3' end!
It took nearly four years from a PhD candidate which culminated in this fantastic JACS paper.
The paper describes how we utilize a special DNA polymerase (TdT) fused to a protein of interest to perform proximity-induced selections. When a DEL member binds to the protein of interest, the proximity to the polymerase causes the latter to extend the DNA strand untemplated. In our method, we only use dATP, and therefore create a pool of binders with a poly-dA tail and a pool of non-binders without it. By using commercially available beads with a poly-dT oligonucleotide, we then can selectively fish out binders - and we do that independently of the binding affinity, as long as the tail surpasses the threshold of the bead step (which is around 20 to 25 nucleotides).
We describe the discovery from a simple model system where we use DNA-DNA affinity as an emulation of protein-DEL member interactions and make the system gradually more complex, from model libraries to a real DNA encoded library. The screen performs well across all conditions and mainly finds the expected hits - with only one false positive motif and an unexpected methoxysulfonamide, which we latter showed that it actually does bind CAII very weakly.
The authors can be proud of themselves. Congratulations, especially to Lukas Schneider who did most of the work!
